Publications
Direct Reprogramming of Fibroblasts to NK Cells
July 15, 2022
MSc. Thesis - University of Porto
Diogo Filipe Pértiga Cabral
Resources:
Abstract
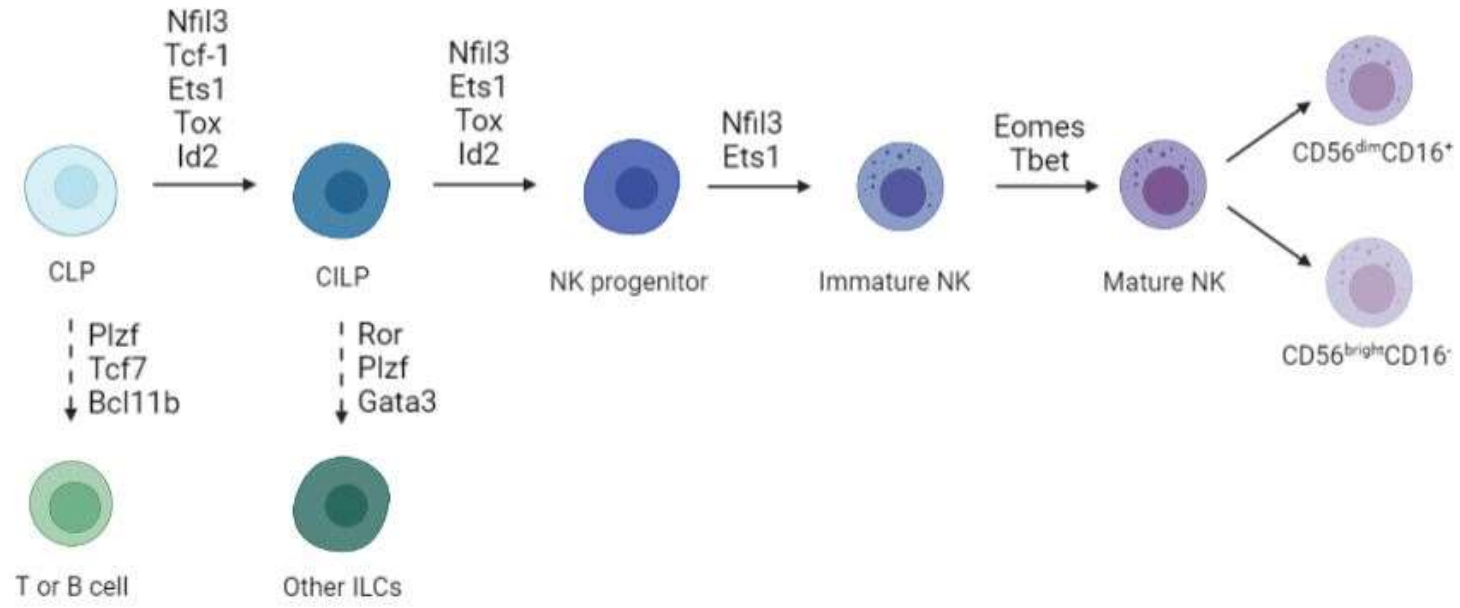
Natural killer (NK) cells are innate lymphocytes with remarkable cytotoxic abilities that control cancer and viral infections independent of antigen specificity. Indeed, NK cells are the first induced pluripotent stem cell (iPSCs)-derived hematopoietic cells to be tested in clinical trials against hematological tumors. However, limited persistence in vivo and complexity of differentiation protocols, pose significant obstacles to widespread NK-based therapeutics. We hypothesize that direct cell reprogramming mediated by cell type-specific transcription factors (TFs) can be employed to generate NK lymphocytes from somatic cells. To define combinations of TF that induce NK cell identity we tested a list of 19 candidate TFs for their ability to activate a NK-specific reporter in mouse embryonic fibroblasts (MEFs). We identified Ets1, Nfil3, T-bet, Eomes (TENE) that activated a NCR1-driven reporter and induce NK progenitor and mature NK global gene expression programs as assessed by single-cell mRNA seq. To evaluate species conservation, we transduced human fibroblasts with 4 TFs and assessed activation of NK-specific markers by flow cytometry. We show that CD34 and CD56 expression is induced by enforced expression of TENE, suggesting that this combination of TFs to induce NK cell identity are conserved in human. CD34 expression increased with time in culture and was enhanced by the addition of cytokines important for lymphocyte development, suggesting cell expansion. Finally, we employed a barcoded TF approach coupled with single-cell mRNA-seq to inform the specification of progenitor versus mature programs. We screened 48 TFs and first confirmed the involvement of Ets1 in immature NK development, T-bet and Eomes in mature NK, and Nfil3 in both stages. We also suggest Runx family TFs and Ikzf1 as novel
regulators of NK development.
Taken together, our results contribute to the understanding of the transcriptional network driving NK lineage commitment and pave the way for the generation of patient- specific NK cells by direct cell reprogramming approaches.